Biewer Terrier Data Analysis Project Update
Project Aims:
• Find out if there are any Biewer Terrier chromosomes which cannot be explained by descent from the Yorkshire Terrier.
• Find other potential breeds which may have influenced the creation of this breed.
Introduction to previous work:
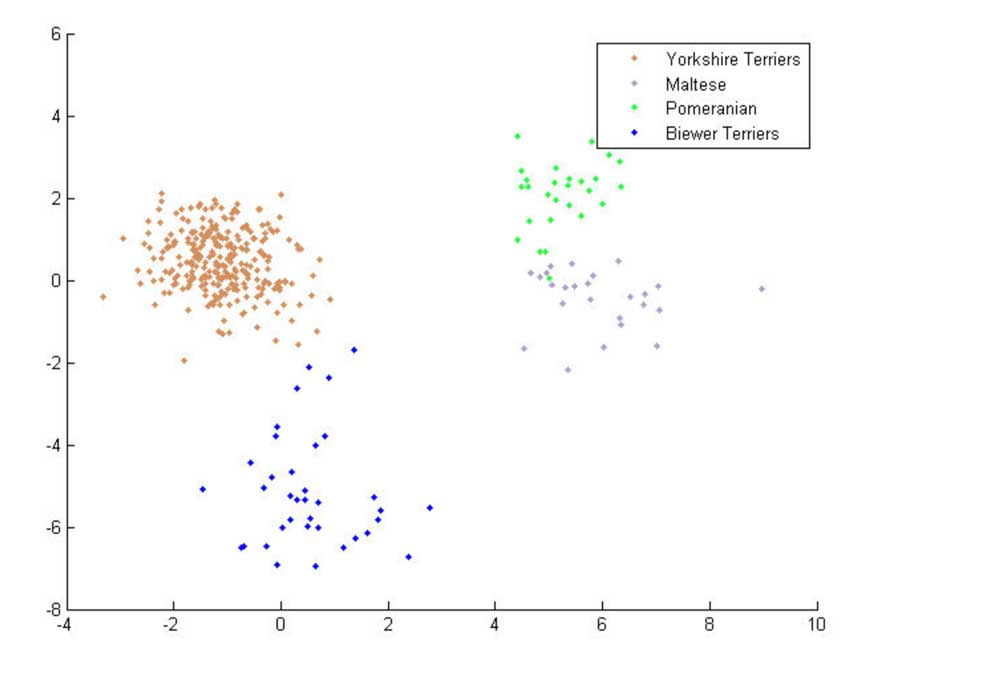
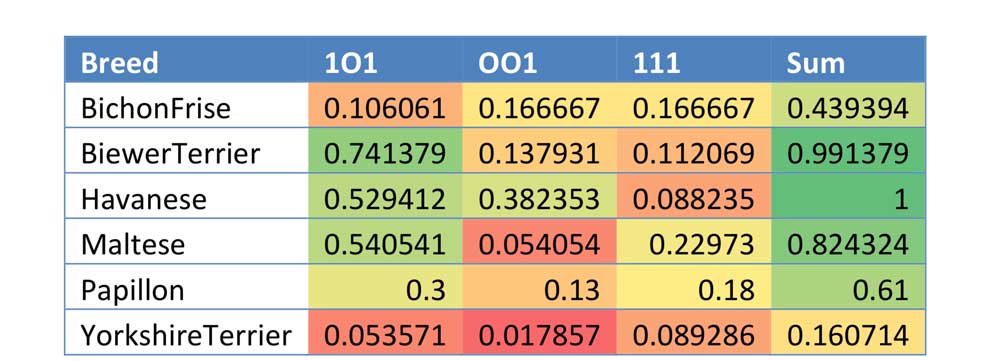
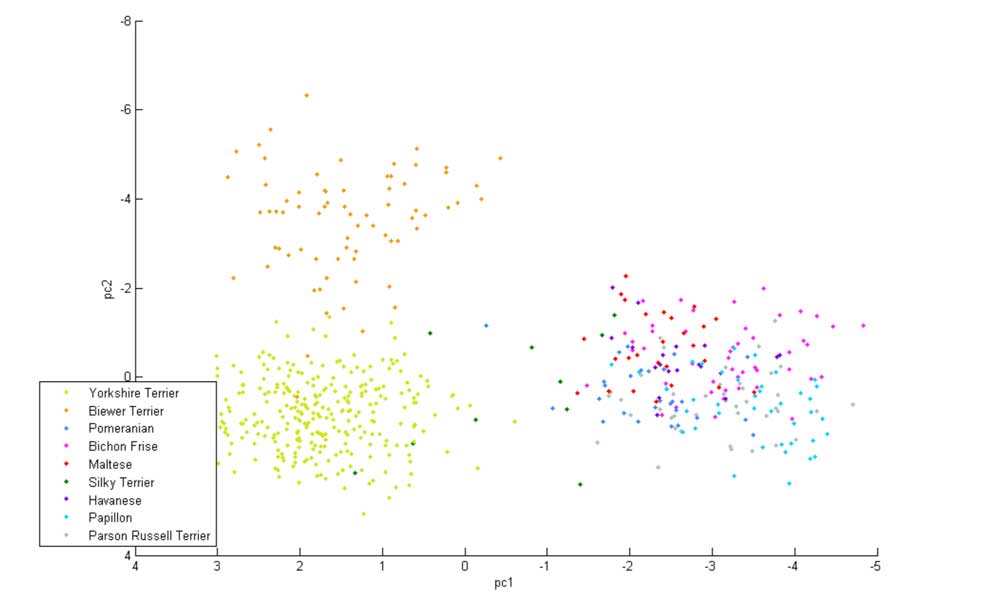
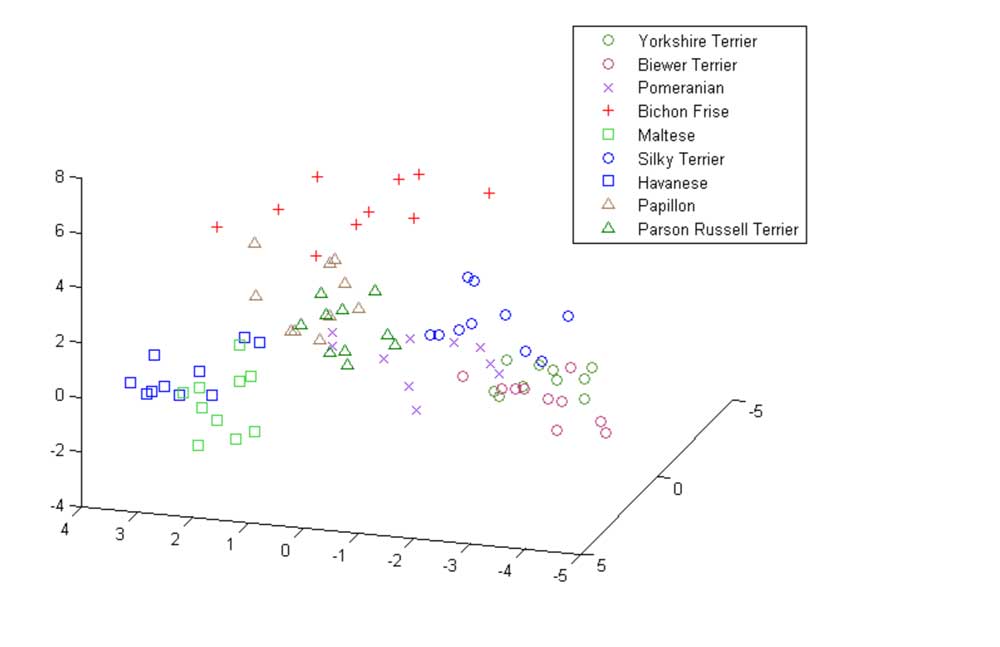
The Biewer Terrier analysis performed so far has encompassed a variety of approaches. The most simple of these used principle component analysis (PCA) to demonstrate that the Biewer Terriers can be clearly distinguished from Yorkshire Terriers using the Wisdom panel MX test SNP data alone, signifying that these dogs have genetic differences and cluster separately from the Yorkshire terriers in the Mars Veterinary database (see Figures 1 ‐ 3).
Figure 1: 3 ‐ Dimensional PCA plot of tested Biewer Terriers and Yorkshire Terriers
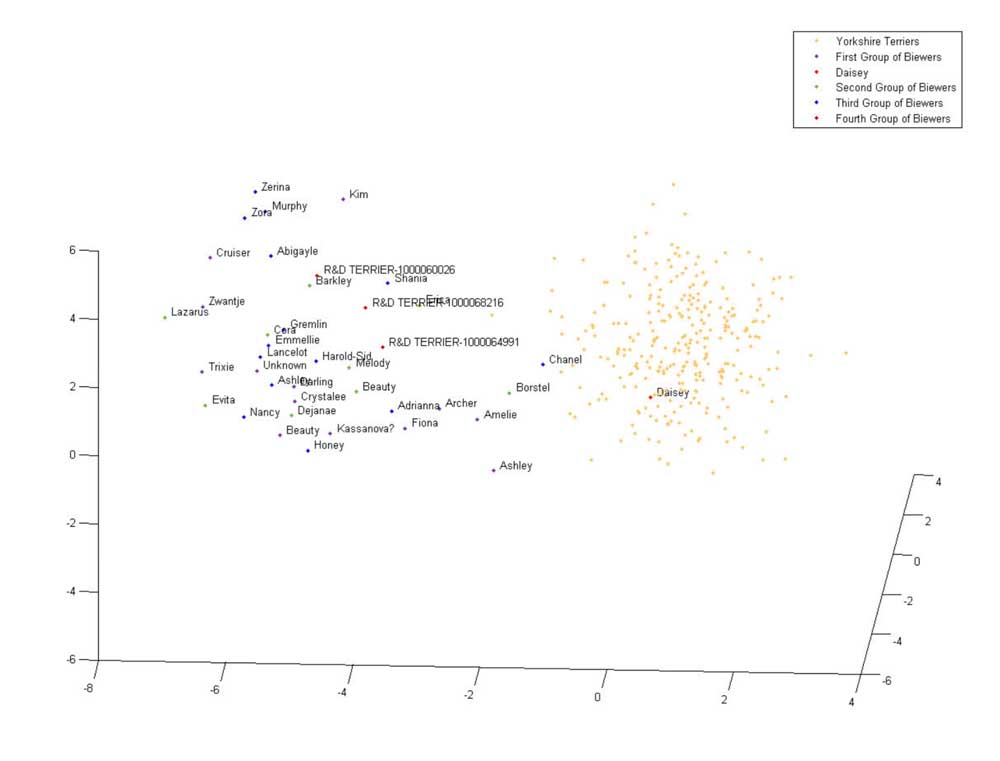
Figure 2: 3 ‐ Dimensional PCA plot of Biewer Terriers and five other breeds
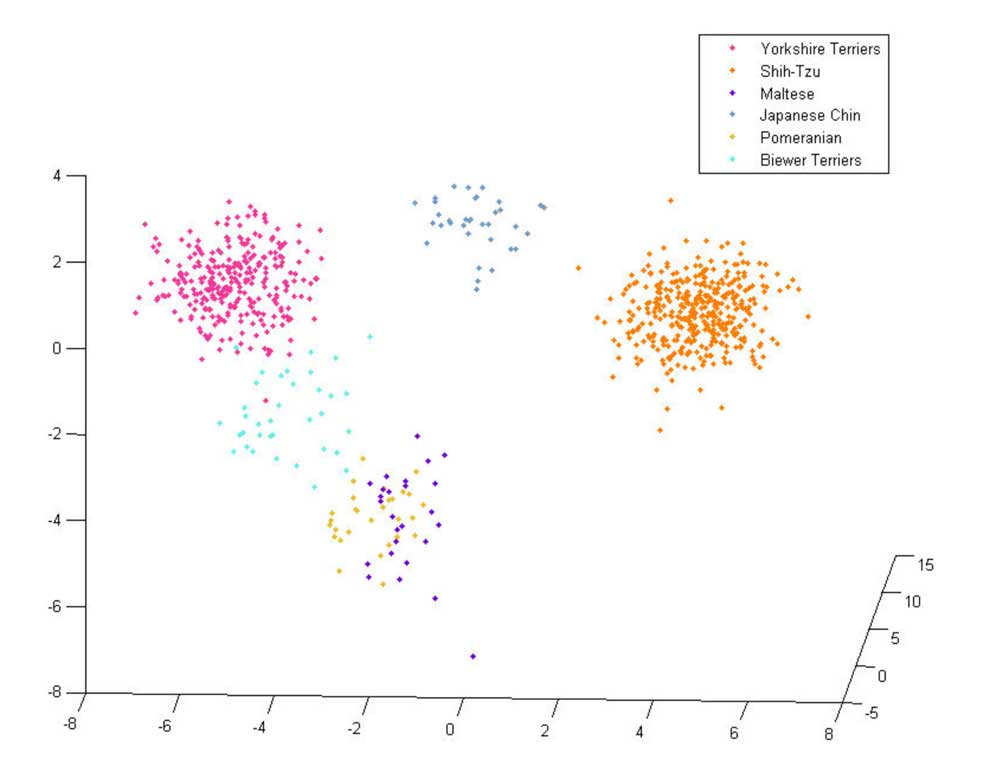
Figure 3: 2 ‐ Dimensional PCA plot showing Biewer Terriers, Yorkshire Terriers, Pomeranians and Maltese breeds
Further analysis:
This document details the other analysis methods that have been used to study the Biewer Terriers in more detail to understand exactly how this breed differs at the genome level from other breeds. The methods employed have included allele frequency analysis and haplotype analysis, including the modeling of mutation and crossover events. We also used PCA analysis on individual chromosomes and with even numbers of breeds. To start with, the calculated distance from breeds in the Mars Veterinary database to the average allele frequencies of the Biewer Terrier was compared to find the breeds which were closest to the Biewer Terrier (data not shown due to size and complexity of spreadsheet). These distances were sorted to find the closest breeds on each of the 25 Chromosomes studied. The most closely related breed at each chromosome was generally the Yorkshire Terrier, though some other breeds were closest to the observed Biewer Terrier average allele frequencies in some places. This served as a quick way to simplify the remaining analysis by allowing the concentration of further efforts on specific genomic areas and run more detailed analysis on less breeds, meaning we could run programs which take a long amount of time, such as PHASE. Nine breeds were selected from the initial comparison of allele frequencies for further detailed analysis and comparison of observed genetic data: Biewer Terriers, Yorkshire Terriers, Silky Terriers, Papillon, Pomeranian, Havanese, Parson Russell Terrier, Bichon Frise, and Maltese.
Quality control of database SNP data:
We performed quality control on the genetic SNP data for individual dogs in our database from the 9 selected breeds, removing SNPs which had observed levels of more than 5% missing data at the locus. We also removed samples which had more than 5% missing data, leaving 162 SNPs for further analysis. This was required because there was some variability in the call success rates of particular SNPs with the different historical analysis methodologies used to generate the SNP data, and also in the DNA quality of samples between different genotyping batches. We attempted to use Phase analysis (see below) to imply the missing data but when we attempted analyzing the Biewer Terrier haplotypes for Yorkshire Terrier haplotypes, they did not exist on Chromosome 20. This was due to missing data near the start of our window of SNPs on this chromosome. Removing just four missing SNPs in this location and repeating phase analysis revealed the presence of Yorkshire Terrier. The other quality controls increased the proportion but at the loss of moving the haplotypes away from the start of this window.
Haplotype analysis using Phase:
Phase uses analysis of SNPs to predict the sequence on the chromosome inherited from each parent. We can use this method to deduce unique sequence haplotypes within a breed population and predict that if other breeds share these haplotypes it is more likely that they share similar chromosomes and a common ancestor.
Haplotype changes reflect how the chromosome changes through generations, the two main disruptive effects are mutation and crossover. Mutation usually alters a single SNP variant within the haplotype and is usually a rare event, and crossover is where the two chromosomes are combined and separated to become a mixture of two ancestor chromosomes. Chromosomal crossover is a common mutational event because it happens during every round of germline sperm and egg cell production by a process called meiosis. Both these types of changes increase the diversity within a breed, though these effects make it more difficult for us to find this new sequence within a different breed.
Using this knowledge of genetics, there were three types of analysis we looked at regarding the haplotypes imputed by phase. The first was looking at which breed populations shared haplotypes with the Biewer Terrier. To find potentially shared ancestry hidden by haplotype changes, we also looked at the likelihood that a Biewer Terrier haplotype could be formed from either the mutation of a haplotype in a separate breed or alternatively from a single chromosomal crossover event.
Shared haplotypes between breeds:
Shared haplotypes were analyzed by tabulating the count of observed haplotypes in each breed. These numbers were divided by the count of all the haplotypes in the breed to get a haplotype frequency.
There are two ways to combine this information. The first is by summing the haplotype frequencies for all the haplotypes of a breed which exist in Biewer Terriers. This gives a metric which portrays how easily those haplotypes would turn up in the Biewer Terriers if that breed had been present in some part of the Biewer Terrier ancestral breed history.
The second method is to sum the haplotype frequency for Biewer Terrier for all non zero haplotype frequencies of a separate breed. This gives an indication of what proportion of the Biewer Terrier population may be explained by the haplotypes of this other breed.
Crossover modeling:
Crossover may be analyzed by creating a list of potential crossovers out of the haplotypes of a population (breed) and counting how many of these may be generated for each haplotype being considered for a chromosome. This gives a likelihood of how easy it is to make this haplotype out of a given population.
For breed populations with large amounts of possible haplotypes, such as Yorkshire Terrier, these numbers were generally small. This suggests that this metric may not be directly compared between breeds. If this is the case, a Boolean approach may be more useful.
Mutation was analyzed in a similar manner to Crossover but by creating a list of potential mutations instead of potential crossovers.
Results:
The overlap of observed haplotype sharing between the other tested breeds and the Biewer Terrier varied quite a lot between chromosomes, as can be seen in Chart 1. There are a large amount of shared haplotypes on Chromosomes 15 and 1, Chromosome 15 is known to harbour the IGF1 gene that is an important determinant of canine size and it is likely that chromosome 1 shares an additional size influencing gene. Chromosome 20 appears to stand out as an enigmatic mystery, with a very small quantity of haplotypes shared with Biewer Terrier from other breeds. If we look at the sum of the combination of haplotype frequencies for each breed, we get a distinct increase in Yorkshire Terrier and a slightly smaller amount spread across Bichon ‐ family type breeds: Havanese, Maltese and Bichon Frise, as if there were some relevant relative of the three which was not in the analysis.
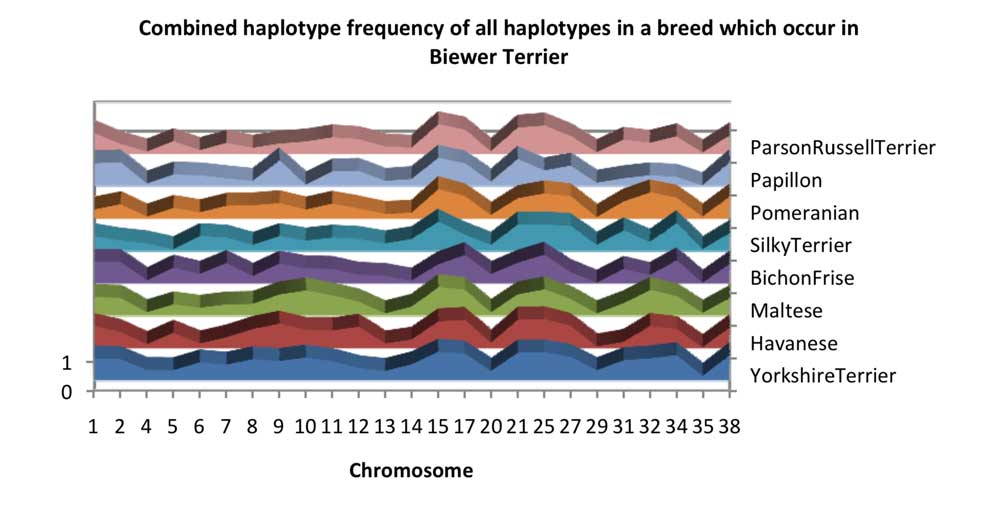
Chart 2 shows the proportion of Biewer Terrier haplotypes which can be explained by their observed presence another breed. It clearly shows how haplotypes which are observed to occur in Yorkshire Terrier could account for a large quantity of the haplotypes which occur in Biewer Terriers. This is perhaps not surprising given the observed physical and genetic similarity of these breeds. Again – chromosome 20 stands out as an obvious exception.
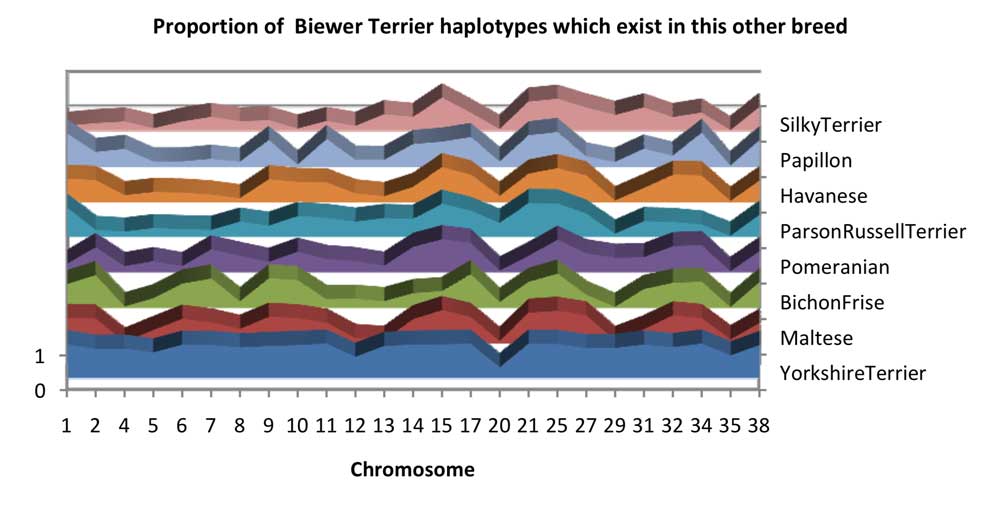
Chart 3 shows the combined likelihood of the Biewer Terrier observed haplotypes being created by a single crossover mutation of haplotypes from the other breeds. This has a lower likelihood in Biewer Terrier for some chromosomes which suggests that these haplotypes may have been created by mutation or directly from other breeds. Chromosome 35 is a good candidate of this, though this chromosome has a larger amount of diversity than the other chromosomes, making this a difficult region to derive accurate lineage information for. There is also a large influence from Bichon ‐ family type breeds such as the Bichon Frise, Maltese, and Havanese.
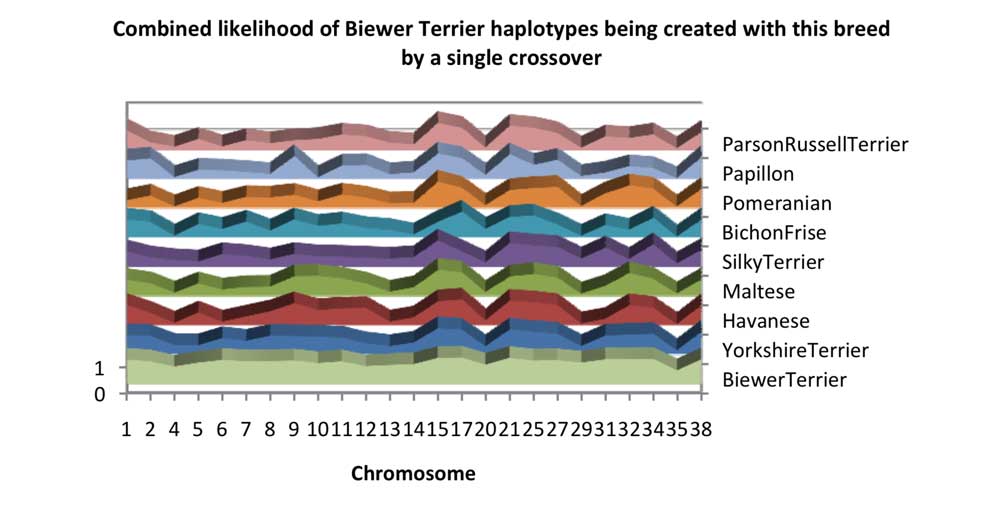
Chart 4 shows the proportion of Biewer Terrier haplotypes which are possible to make with a crossover of two haplotypes of a different breed. Almost all the haplotypes are possible if you had Yorkshire Terrier population and used a single crossover. Inheritance of Yorkshire Terrier breed haplotypes can explain the haplotype variety seen in the Biewer Terrier if crossover mutations are considered, even on Chromosome 35. This is not 100% true for the other breeds.
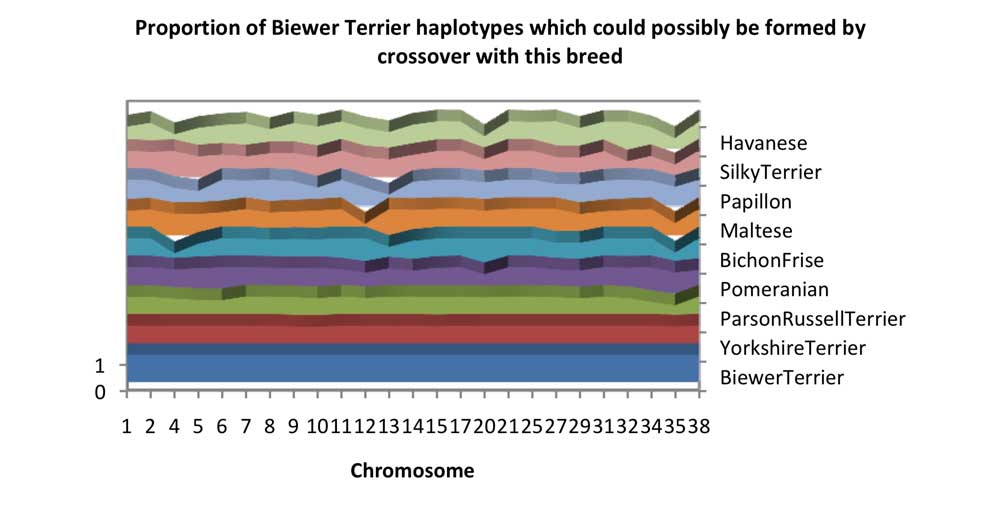 Chart 5 looks at the combined likelihood of a SNP mutation resulting in Biewer Terrier haplotypes. The observed pattern is similar between most of the breeds.
Chart 5 looks at the combined likelihood of a SNP mutation resulting in Biewer Terrier haplotypes. The observed pattern is similar between most of the breeds.
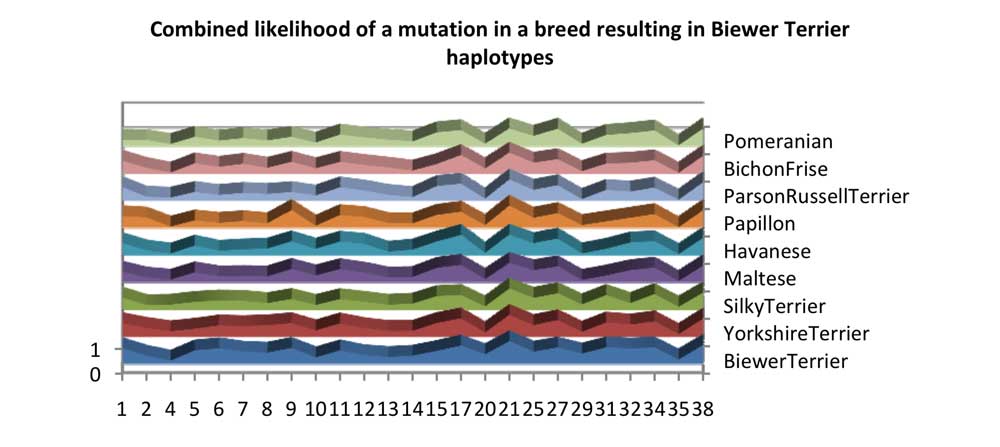
Chart 6 shows the proportion of Biewer Terrier haplotypes which can be explained with mutations of other breeds. Some breeds have a larger effect as they share the haplotypes which may be mutated to create other Biewer Terrier haplotypes. Chromosome 4 looks odd, but the most common Biewer Terrier haplotype has a haplotype frequency of 33%, and can only be created from crossover within this population.
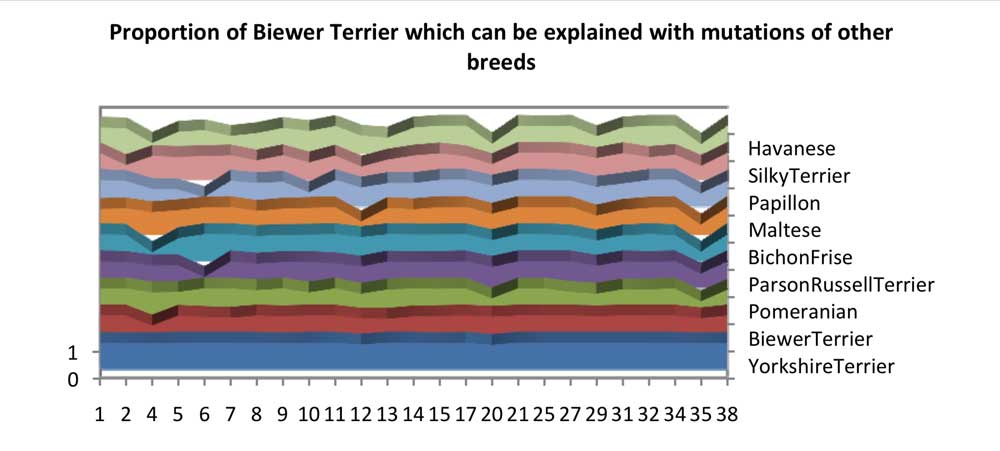
Investigation of Chromosome 20 haplotypes:
To summarize the data presented so far, we have detected a strong Yorkshire Terrier influence on all chromosomes except chromosome 20. We now address the Chromosome 20 enigma with further analysis of this Chromosome.
Chart 7 shows the ranks of haplotype frequency in Biewer Terrier on Chromosome 20. The larger the bar, the higher the observed haplotype ranked haplotype frequency in this breed. Chart 7 shows that although Yorkshire Terrier does not share every haplotype with Biewer Terriers, the haplotypes that it does share are some of the most common haplotypes in the Biewer Terrier breed, including three haplotypes which have the top three ranked haplotype frequencies in the Yorkshire Terrier set of haplotypes. On this chart, there is the observation that Parson Russell Terrier is the only breed sharing the most common Biewer Terrier haplotype. This haplotype has an observed haplotype frequency of 34% in Biewer Terrier, yet only 0.02% in Parson Russell Terriers, so it is unlikely to be a good hit and is probably unlikely to explain the origin of this haplotype in Biewer Terriers.
Chart 7: Ranks of how common each haplotype in Biewer Terrier is for each of the breeds on Chromosome 20. 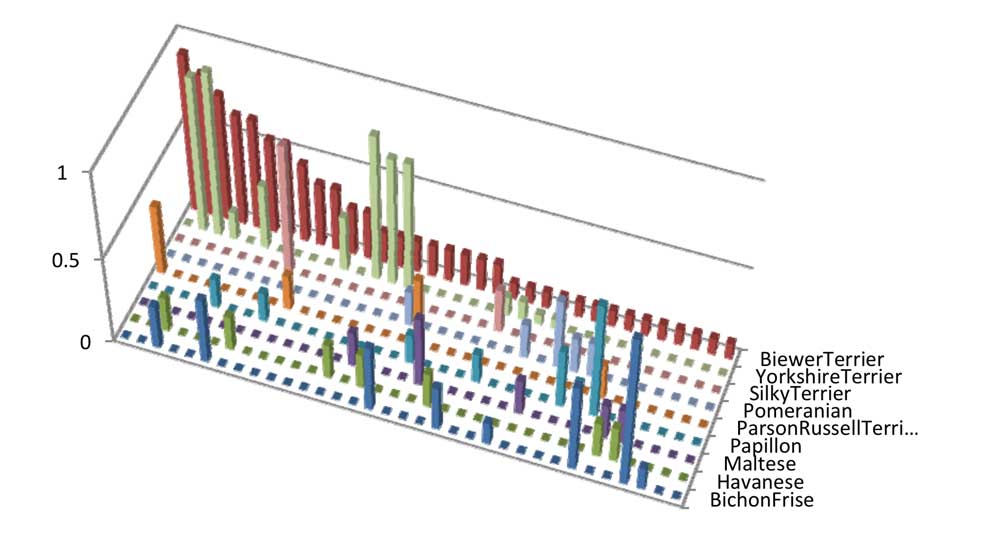
The most common haplotype in Biewer Terriers for chromosome 20 has such a high haplotype frequency (34% of all Biewer Terrier haplotypes), yet does not seem to share this haplotype with other breeds supported with a reasonable haplotype frequency. This indicates that this haplotype may be extremely important for determining a key character trait in Biewer Terriers.
Looking further into this region for any relevance to the Biewer Terrier we found a gene which is possibly the most relevant to the Biewer Terrier’s characteristic coat and pattern named MITF. MITF is a gene which has been associated with regulating white patterning in dogs, and in essence, says where the dog will have pigmented cells that produce any coat color. The absence of this gene product in cells makes regions of a dog’s coat white.
The location of this gene is within the start of the first few SNPs the Wisdom Panel database uses on Chromosome 20, which was one of the regions where we removed SNPs for the previous experiments due to stringent quality control measures.
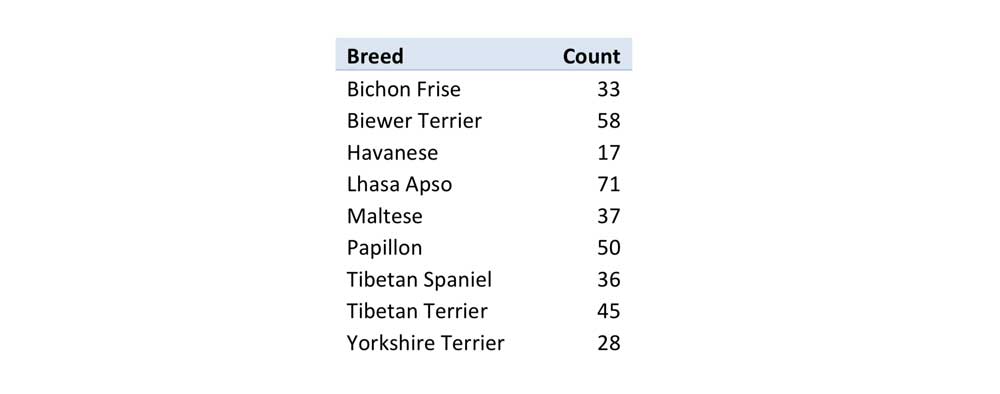
To investigate this region more fully, we focussed on the investigation of haplotypes from samples which had no missing information for the first 6 SNPs of Chromosome 20. The count of the samples analyzed for these SNPs are shown in table 1. The haplotype frequencies of these haplotypes determined by Phase using these 6 SNPs are shown in Chart 8. Chart 8 shows that Havanese shares 6 out of the 13 observed Biewer Terrier haplotypes. There were only three other haplotypes identified in Havanese which did not occur in Biewer Terriers. This makes Havanese a good match for the Biewer terrier haplotypes found in this region. The Bichon Frise haplotypes seem to be shared between either Havanese or Maltese. The second most common Biewer Terrier haplotype is one which is seen in Papillon and to a lesser extent in Yorkshire Terriers, and this haplotype could potentially create the most common haplotype found in Biewer Terriers through a single mutation event.
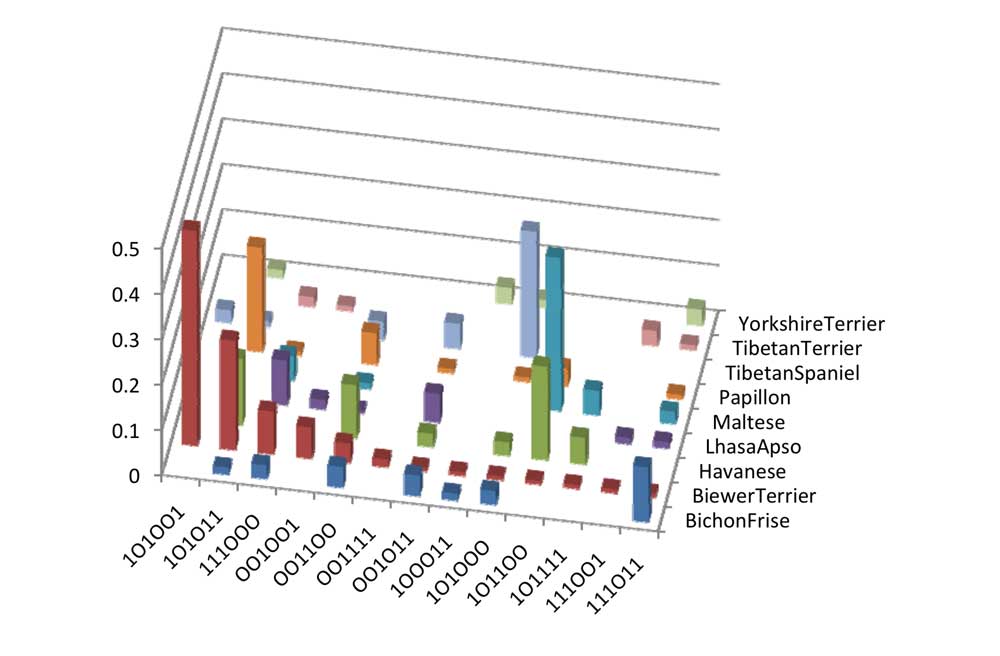
Table 1: Quantity of dogs used for investigating Chromosome 20’s MITF region.
If we focus even closer to the MITF gene by looking at the three most common haplotypes of the first three SNPs in Biewer Terriers we get Table 2. This shows that these three sequences are the only ones which exist in the 17 selected Havanese samples. The haplotype frequency for these haplotypes is low in Yorkshire Terrier, but not non ‐ existent. This implies that these haplotypes would be much easier to inherit from Havanese dogs than Yorkshire Terriers. If this haplotype is associated with the Biewer coat color, it also suggests that it is more likely to have come from a Havanese ‐ family type breed rather than Yorkshire Terrier.
Table 2: Combined haplotype frequency of first three SNPs of Chromosome 20.
Further PCA analysis:
Principle component analysis was run in Matlab on the chromosome 20 data set (which had the average allele frequencies for the SNP substituted for any missing SNP data). This resulted in the plot shown in chart 9. This treats Yorkshire Terrier and Biewer as distinct breeds, yet merges all the other breeds. This is due to the analysis using a disparate amount of samples.
Chart 9: PCA of genotypes for selected samples.
If we equalize the number of samples given to each breed by selecting ten samples for each breed at random, we get the plot in chart 10. In this case, there are three clusters of breeds. There is a Maltese / Havanese cluster, a Papillon / Parson Russell Terrier cluster, and a Biewer Terrier / Yorkshire Terrier / Silky Terrier cluster. The Biewer Terrier is quite far away from the potential Havanese influence on this plot, though it clusters quite closely to the Yorkshire Terrier. This could possibly be a further disparate number of samples effect due to the Silky Terrier adding to the Yorkshire Terrier influence, or the inclusion of the sole Asian breed, the Pomeranian.
Chart 10: PCA of genotypes for 10 randomly selected samples of these breeds.
Conclusions:
Biewer Terriers share the majority of their haplotypes with Yorkshire Terriers.
Chromosome 20 is a less perfect match to Yorkshire Terrier. The region where these two breeds are most different on Chromosome 20 is around the MITF region, which is known to be related to white coat patterning. The closest breed at this location that we have data for is the Havanese, though the other Bichon Frise family breeds are relatively close as well.
Current Position:
• Find out if there are any Biewer Terrier chromosomes which cannot be explained by descent from the Yorkshire Terrier. Yes, chromosome 20 looks different and has a region where Yorkshire Terrier is different to the Biewer Terrier.
• Find other potential breeds which may have influenced the creation of this breed. The closest breed to the Biewer Terrier where Yorkshire Terrier is most different is the Havanese.
Information on the MITF region:
We have looked at a recent paper on MITF in dogs, and it highlights two relevant genetic variants which are related to different forms of white coat patterning. The first is a SINE insertion, and the second is a length polymorphism.
It is possible to do experiments to find the presence of the SINE insertion. Some breeds with white patterning contain this variant, whereas the Yorkshire Terriers that were tested in the paper do not. If Biewer Terriers are observed to have this insertion in a similar location to other breeds, we will know that Biewer Terriers have another breed in its makeup as the presence of a de ‐ novo insertion in this region in an ancestral Yorkshire Terrier is highly unlikely. The absence of this insertion in Biewer Terriers would not tell us anything significant.
We could also potentially sequence the length polymorphism. This would give us data on a short sequence of DNA which is unique between white spotted and non-spotted dogs. We should be able to tell if it is more likely that the Biewer Terrier copy of this sequence is genetically closest to Yorkshire Terrier, or an alternate breed if data on enough breeds in this region were determined. This is potentially a challenging task, as this length polymorphism contains a form of the sequence which is typically problematic for sequence analysis.
As of date, both experiments we have attempted to look at these variants in the laboratory have failed, though trying different primers and experiments could potentially resolve the current issues.
